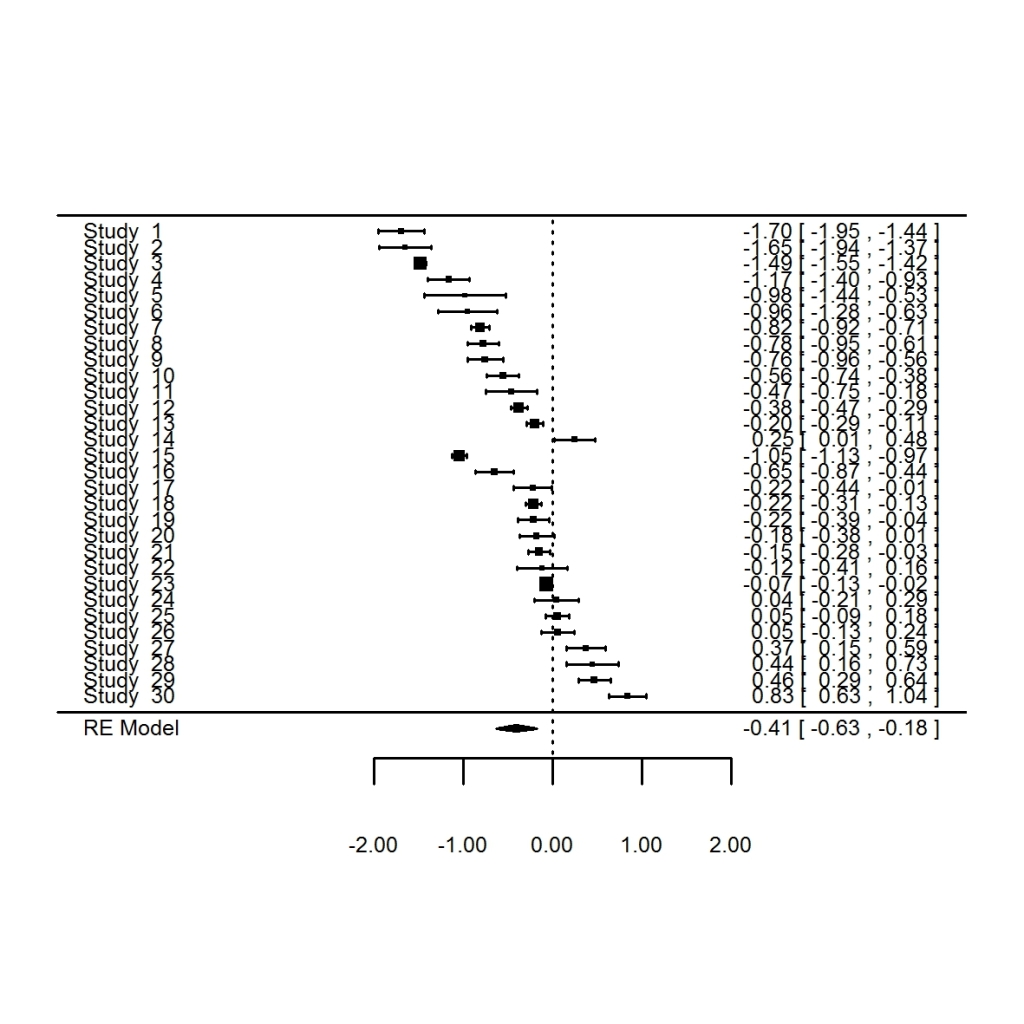
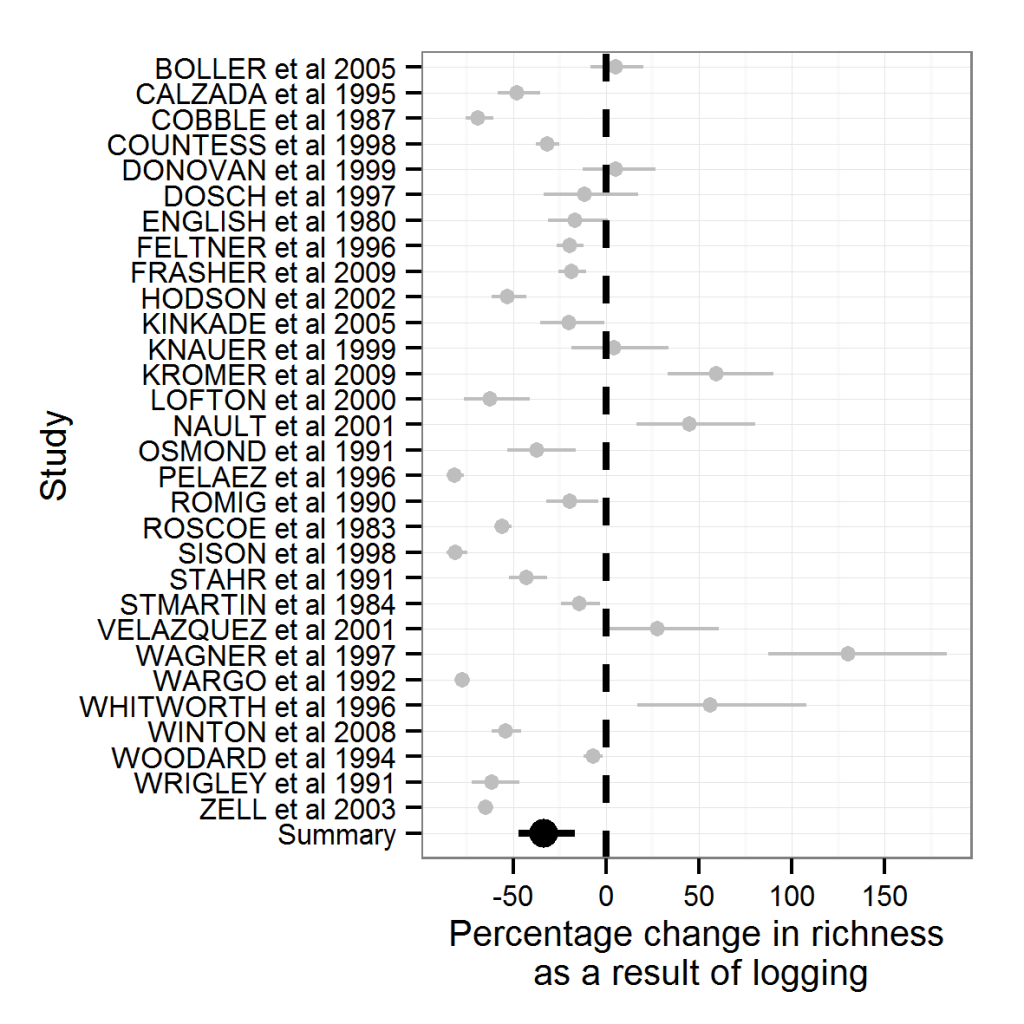
In the last post I introduced you to the basics of meta-analysis using metafor together with ggplot2. If you haven’t seen that post, I suggest you go back and read it since this post will be hard to follow otherwise.
In this post I will concentrate on trying to explain differences between different sites/studies.
Going back to our previous example, we saw that logging tended to have a negative effect on species richness.
Luckily my completely made up data had information on the intensity of logging (volume of wood removed per hectare) and the method used (conventional or reduced impact).
We can make a model looking at the effects of logging intensity and method on species richness. First we can start with our most complex model
Model1<-rma.uni(yi,vi,mods=~Intensity*Method,method="ML",data=ROM) summary(Model1)
This spits out the following:
Mixed-Effects Model (k = 30; tau^2 estimator: ML)
logLik deviance AIC BIC
-15.8430 121.7556 41.6860 48.6920
tau^2 (estimated amount of residual heterogeneity): 0.1576 (SE = 0.0436)
tau (square root of estimated tau^2 value): 0.3970
I^2 (residual heterogeneity / unaccounted variability): 96.52%
H^2 (unaccounted variability / sampling variability): 28.74
Test for Residual Heterogeneity:
QE(df = 26) = 1151.1656, p-val &lt; .0001
Test of Moderators (coefficient(s) 2,3,4):
QM(df = 3) = 38.2110, p-val &lt; .0001
Model Results:
estimate se zval pval ci.lb ci.ub
intrcpt 0.1430 0.3566 0.4011 0.6883 -0.5558 0.8419
Intensity -0.0281 0.0099 -2.8549 0.0043 -0.0474 -0.0088 **
MethodRIL 0.1715 0.4210 0.4074 0.6837 -0.6536 0.9966
Intensity:MethodRIL 0.0106 0.0138 0.7638 0.4450 -0.0166 0.0377
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Looking at our residual plots, they seem to conform to our assumptions of constant variance.
plot(fitted.rma(Model1),residuals.rma(Model1)) qqnorm.rma.uni(Model1)
Our models suggests that the more wood is extracted from a forest the greater the negative effect on species richness, but that seems to be little evidence about the effect of method or the the interaction between intensity and method.
So, lets get rid of the interaction and see what happens:
Model2<-rma.uni(yi,vi,mods=~Intensity+Method,method="ML",data=ROM) summary(Model2)
Mixed-Effects Model (k = 30; tau^2 estimator: ML)
logLik deviance AIC BIC
-16.1323 122.3341 40.2646 45.8694
tau^2 (estimated amount of residual heterogeneity): 0.1606 (SE = 0.0443)
tau (square root of estimated tau^2 value): 0.4008
I^2 (residual heterogeneity / unaccounted variability): 96.75%
H^2 (unaccounted variability / sampling variability): 30.80
Test for Residual Heterogeneity:
QE(df = 27) = 1151.4310, p-val &lt; .0001
Test of Moderators (coefficient(s) 2,3):
QM(df = 2) = 36.9762, p-val &lt; .0001
Model Results:
estimate se zval pval ci.lb ci.ub
intrcpt -0.0416 0.2648 -0.1570 0.8753 -0.5606 0.4774
Intensity -0.0228 0.0070 -3.2617 0.0011 -0.0364 -0.0091 **
MethodRIL 0.4630 0.1803 2.5681 0.0102 0.1096 0.8163 *
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Now both intensity and method are statistically significant, but method less so. Let’s take it out:
Model3<-rma.uni(yi,vi,mods=~Intensity,method="ML",data=ROM) summary(Model3)
Mixed-Effects Model (k = 30; tau^2 estimator: ML)
logLik deviance AIC BIC
-19.1526 128.3747 44.3052 48.5088
tau^2 (estimated amount of residual heterogeneity): 0.1962 (SE = 0.0536)
tau (square root of estimated tau^2 value): 0.4429
I^2 (residual heterogeneity / unaccounted variability): 97.45%
H^2 (unaccounted variability / sampling variability): 39.27
Test for Residual Heterogeneity:
QE(df = 28) = 1258.6907, p-val &lt; .0001
Test of Moderators (coefficient(s) 2):
QM(df = 1) = 25.2163, p-val &lt; .0001
Model Results:
estimate se zval pval ci.lb ci.ub
intrcpt 0.4678 0.1927 2.4276 0.0152 0.0901 0.8455 *
Intensity -0.0324 0.0065 -5.0216 &lt;.0001 -0.0451 -0.0198 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Unsurprisingly, intensity is now more significant. However, we have taken out method which may have some value in explaining differences. The tool I use most for model selection is AIC. This basically aims to get the best fitting model with the least explanatory variables. This is why I did the last step of removing method. It could be that the effect of method is significant but adds relatively little explanatory power to our model.
Let’s test this by calculating their AICs.
AIC(Model1) AIC(Model2) AIC(Model3)
[1] 41.68604 > AIC(Model2) [1] 40.26459 > AIC(Model3) [1] 44.30516
We choose the model with the lowest AIC, in this case Model2.
To calculate the fit of our model, we can compare it to our intercept only model (the original meta-analysis from part 1), but we have to set method=”ML” since REML is a poor estimator of deviance.
ROM.ma2&lt;-rma.uni(yi,vi,method=&quot;ML&quot;,data=ROM) summary(Model2) summary(ROM.ma2)
Using the two Tau squared statistics we can calculate the proportion of deviance not explained by the intercept only model using the sum:
in.plot<-ggplot(data=ROM,aes(x=Intensity,y=yi,size=1/vi,colour=Method))+geom_point(data=ROM,shape=16)
in.plot2<-in.plot+geom_line(data=ROM,aes(x=Intensity,y=preds,size=1))
in.plot3<-in.plot2+ylab("log response ratio")+xlab("Logging intensity (m-3 ha-1)")
in.plot3+theme(legend.position="none")
Circle size is relative to weight for each study, since we weight the most precise studies most heavily. Blue points and line refer to reduced impact logging sites, while red refers to conventionally logged sites.
We can also transform it all to proportions for more intuitive interpretations:
theme_set(theme_bw(base_size=10))
in.plot<-ggplot(data=ROM,aes(x=Intensity,y=exp(yi)-1,size=1/vi,colour=Method))+geom_point(data=ROM,shape=16)
in.plot2<-in.plot+geom_line(data=ROM,aes(x=Intensity,y=exp(preds)-1),size=1)
in.plot3<-in.plot2+ylab("Proportional change\nin species richness")+xlab("Logging intensity (m-3 ha-1)")
in.plot3+theme(legend.position="none")

The graphs show that, for my made up data, reduced impact logging has a less negative effect than conventional logging on species richness when carried out at similar intensities but that increasing intensity leads to reductions in species richness for both conventional and reduced impact methods.
I think that wraps it up for now. I hope you’ve found this useful. As ever any comments or questions are welcome.
I will be back to writing non-stats posts again soon!